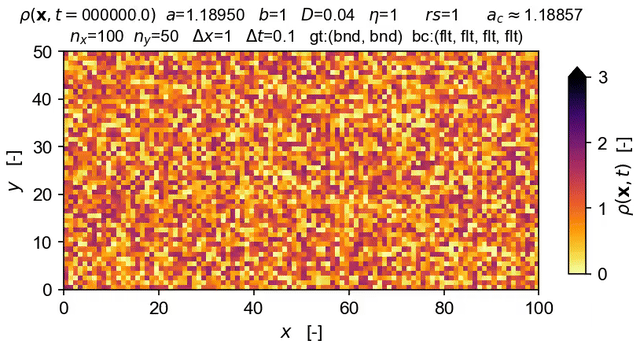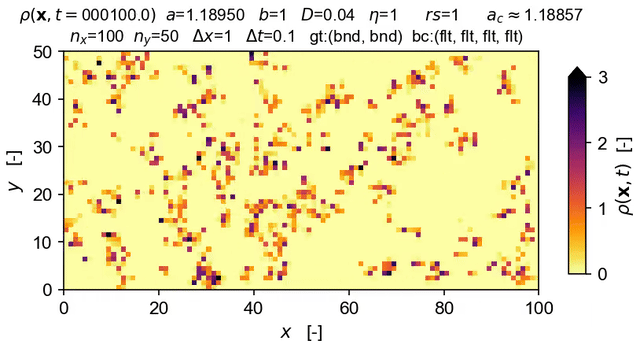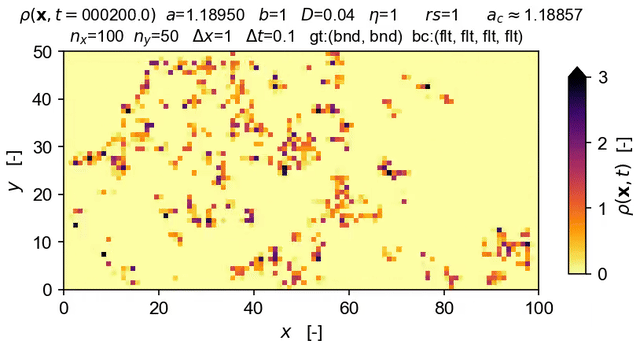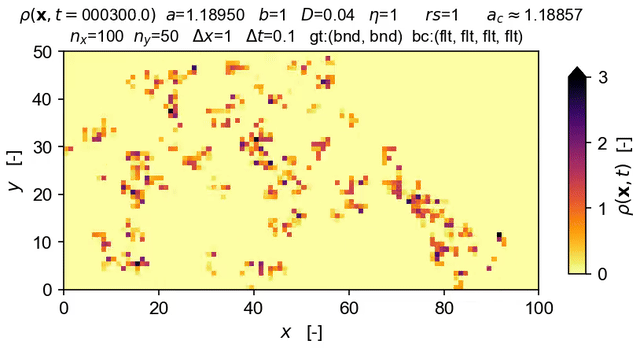
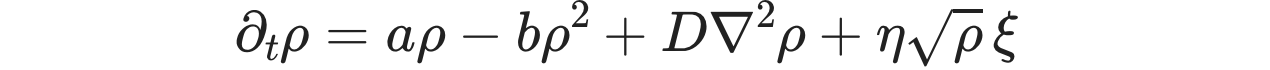Tools to integrate Langevin equations of absorbing phase transition (APT) type — with a focus on solution of the directed percolation (DP) Langevin equation.
The langevin package implements the operator-splitting method originally developed by Dornic et al (2005), Pechenik & Levine (1999) and others, and improved upon by Weissmann et al (2018).
It provides a Python wrapper around core C++ heavily adapted from a code base written by Paula Villa Martín, extended by Victor Buendía ("VMB"), and arising from earlier efforts by Ivan Dornic and Juan Bonachela. The wrapper provides easy access to the Langevin integrator, and broad opportunity to experiment, adapt, and extend it further.
The current C++ implementation extends the VMB code to allow run-time specification of grid dimension and size, boundary topology (bounded or periodic), boundary conditions, and initial conditions. It further provides tools for running model integration in batches, time-slicing the Langevin field grid, and recording of time-series of grid properties.
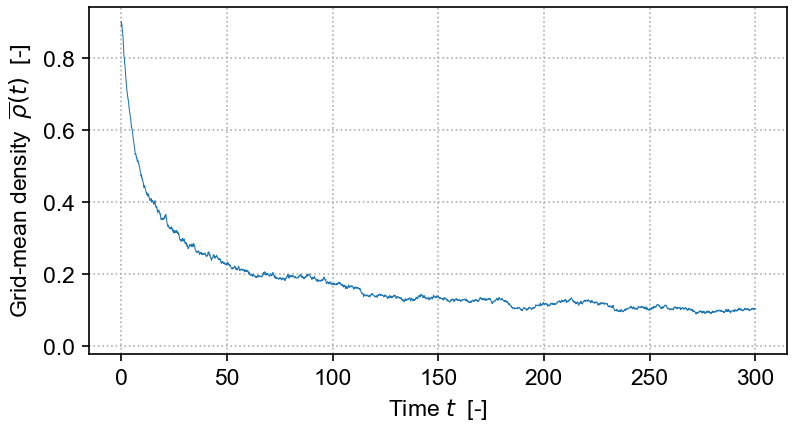
The equation solved in the demo here is the DP Langevin for a 2D grid with initial values sampled from U[0,1]:
where ρ(x,t) is the order parameter field, a and b are rate constants, D is the diffusion rate over x, ξ(x,t) is Gaussian white noise (uncorrelated, zero mean, unit variance), and η is the "demographic" noise amplitude.
See Victor Buendía's fork of Paula Villa Martín's repo for details on more general applications and on how the integration scheme is implemented.
The structure of the DP/APT Langevin-equation integrator package is broadly as follows (detailed documentation is available here).
First, there is a wrapper file called cplusplus/dp/wrapper_dplvn.cpp that uses pybind11 to link the C++ code to a Python runtime.
Next, the code is split into a hierarchy of three groups, with each corresponding file denoted by one of following prefixes: (1) sim_dplangevin_, (2) dplangevin_ and (3) langevin_:
-
The
cplusplus/dp/sim_dplangevin_*files provide aSimDPclass, made available through the wrapper at the Python level, required to manage and execute DP Langevin model integration. ThisSimDPclass instantiates aDPLangevinclass integrator to do the hard work of numerical integration of the stochastic differential equation. Langevin field density grids are returned to Python (via the wrapper) asnumpyarrays as are time series of the mean density field and its corresponding epochs. -
The
cplusplus/dp/dplangevin_*files define thisDPLangevinintegrator class. They inherit the generalBaseLangevinintegrator class and implement several methods left undefined by that parent; most important, they define methods implementing the particular functional form of the directed-percolation Langevin equation and its corresponding nonlinear, deterministic integration step in the split operator scheme.Other types of absorbing-phase transition-type Langevin equation could be implemented with alternate subclasses of
BaseLangevinand alternate versions of theSimDPclass. -
The
cplusplus/langevin_*source files provide the baseBaseLangevinclass that implements the operator-splitting integration method in a fairly general fashion. Grid geometry and topology, boundary conditions, initial conditions, the integration scheme, and a general form of the Langevin equation are all coded here. The core Dornic-style integrator is a heavily altered version of the Villa-Martín and Buendía code.
For here for more comprehensive installation notes that cover multiple platforms. The info below applies only to Linux and macOS.
First, set up a suitable Python environment.
The simplest tool is uv, but there are several other options.
If you use conda or miniconda, take a look at the environment.yml file provided.
We recommend installing Python 3.14 since development of langevin uses this version.
For example, if you're using uv, all that's needed is to create an
appropriately named folder, navigate to it, and execute:
uv venv --python=3.14
source .venv/bin/activate
where the --python option forces uv to choose that version of the Python intepreter.
Then, install the langevin package from PyPI:
pip install langevin
if you're using uv, this command will be
uv pip install langevin
This step should automatically install all the dependencies as well. If it does not, see below.
If you want to access more regular updates, you can install from TestPyPI:
[uv] pip install --index https://test.pypi.org/simple/ \
--default-index https://pypi.org/simple/ langevin
Note: the --default-index ensures that package dependencies are fetched from the main PyPI repository where needed.
At minimum, langevin needs Python≥3.12 and the package pybind11. To run the demos, you will also need numpy, matplotlib, jupyter, ipython, along with pandas, tqdm, and ffmpeg-python.
If you are using conda or miniconda, it would be best to install them using
the environment.yml file, instead of relying on pip to do the job (mixing pip and conda is not a great idea anyway, but langevin is not yet available on conda).
If you want to build locally, you will also need meson-python, wheel, pybind11, and ninja.
To turn density field image sequences into animations, langevin uses ffmpeg-python, which assumes that ffmpeg is itself installed on your system.
On Linux platforms, matplotlib has a tendency to complain about missing fonts, e.g., Arial, generating large numbers of warnings in some of the notebooks. This can be fixed by installing the missing fonts and ensuring that matplotlib's cache is refreshed.
We currently have pre-built binary wheels macOS 14, macOS 15, the latest macOS build, and multiple flavors of Linux (most of which have been tested), as well as Windows (but not yet tested).
If your platform is not explicitly supported with a pre-built binary, the following will force a build from source:
[uv] pip install -v langevin --no-binary langevin
The package can also be built "by hand."
Some build info is provided in the cplusplus/ directory. The build system is meson-python, using pybind11 as the C++ wrapper.
Simple demonstration scripts are provided in demos/. More complete examples are provided in the simulation/ directory. The easiest route is to git clone the repo to get these files, or you can download one-by-one.